Purpose of this post
I enjoy finding new ways to visualize data in a meaningful (and also aesthetically pleasing) way. Finding the right tools in order to get your message across adds value, but can be challenging. When I learned about this visualization toolbox with the multitude of options it has, I had to learn how to use it – and it’s actually quite simple!
The links to both the blog and GitHub pages below are extremely informative (so please also refer to them)! Here, I wanted to outline the installation process I used in order to get the program running in RStudio (Part 1) and provide some examples of things I have done with the toolbox to potentially spark some inspiration (Part 2)!
Getting Started
Source
All the credits for this awesome toolbox go to Dr. Athanasia Mowinckel & Dr. Didac Vidal Pineiro at the Center for Lifespan Changes in Brain and Cognition, University of Oslo. Be sure to check out Dr. Mowinckel’s blog, which has a great overview of the different options available with the toolbox.
ggseg GitHub link: https://github.com/LCBC-UiO/ggseg
ggseg GitHub README.md: https://github.com/LCBC-UiO/ggseg/blob/master/README.md
Note: There seem to be some updates happening at the moment with the toolbox, so this may not work 100% for you, but it worked for me in May of 2020. Updates to come if this changes.
Compatibility – what I used
Operating System: Windows 10
R version: 4.0.0
RStudio version: 1.2.5033
Installation Guide
Libraries to install prior to ggseg
library(ggplot2) library(devtools) library(tidyselect) library(magrittr) library(rmarkdown) library(stringr) library(stringi) library(tibble) library(dplyr) library(purrr) library(pandocfilters) library(tidyr)
If some libraries are missing, use the command below with the toolbox name within the quotes as shown for tidyr.
$> install.packages("tidyr")
Installing ggseg
On the GitHub website, they state to use:
$> install.packages("remotes")
remotes::install_github("LCBC-UiO/ggseg", build_vignettes = TRUE)
Currently, there is a bug that is being fixed in the program with the Vignettes, so you will need to modify the code until this is fixed to:
$> install.packages("remotes")
$> remotes::install_github("LCBC-UiO/ggseg", build_vignettes = FALSE)
Personally, for me, I found that devtools works for installing the program without any problems. So since I already installed devtools above, just enter the following in RStudio:
$> devtools::install_github("LCBC-UiO/ggseg", build_vignettes = FALSE)
$> library(ggseg)
This will take a little bit of time and install version 1.5.4. You should see this in your window as:
$> building 'ggseg_1.5.4.tar.gz'
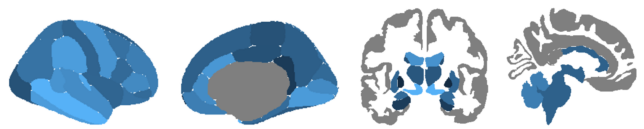
Atlases
After installing ggseg, there are two default atlases pre-installed based on the default Freesurfer output from running recon -all:
- Desikan-Killany cortical atlas (dk)
- Subcortical segmentation atlas (aseg)
Additionally, the following atlases are available and the links show examples for how to use the atlases. These are beyond the scope of this post. I will focus just on the default atlases included in the package.
- Yeo – both 7 and 17 network data
- Desterieux – the 2009 atlas
- Chen – both thickness and area maps
- Schaefer – both 17 and 7 networks
- Glasser – full atlas
- JHU – white tract atlas
- Tracula – white tract atlas
- ICBM – white tract atlas
- HO – Harvard-Oxford cortical (FSL)
- DefaultExtra – extra 2d view for dk, p/a division
To install the extra atlases, first install ggsegExtra:
$> devtools::install_github("LCBC-UiO/ggsegExtra", build_vignettes = FALSE)
$> library(ggsegExtra)
To check to see which atlases are available enter the following command:
$> ggseg_atlas_repos()
Install the atlas you would like to use with the format below (using the Glasser atlas as an example):
$> devtools::install_github("LCBC-UiO/ggsegGlasser")
$> library(ggsegGlasser)
Now you should hopefully have ggseg installed and ready to go without any problems!